Key Highlights of Task 1:
- Mitotic Figures as Key Biomarkers: Mitotic figures are essential for evaluating tumor proliferation and play a critical role in cancer grading.
- Detection Challenges: Their morphology often overlaps with similar structures, leading to variability and frequent false positives.
- Current AI Limitations: Existing algorithms struggle with tissue variability and often fail in non-tumor, inflamed, or necrotic regions.
- Expanded Scope in MIDOG 2025: This task focuses on detecting mitotic figures across all tissue regions, including non-hotspot areas, to ensure real-world applicability.
- Objective: Develop robust, accurate algorithms that minimize false positives and perform consistently across diverse histological regions.
Enhancing Accuracy in Mitotic Figure Detection
The morphology of mitotic figures strongly overlaps with similarly looking imposters. Therefore, the mitotic count is notorious for having a high inter-rater variability, which can severely impact prognostic accuracy and reproducibility. This calls for computer-augmented methods to help pathology experts in rendering a highly consistent and accurate assessment.
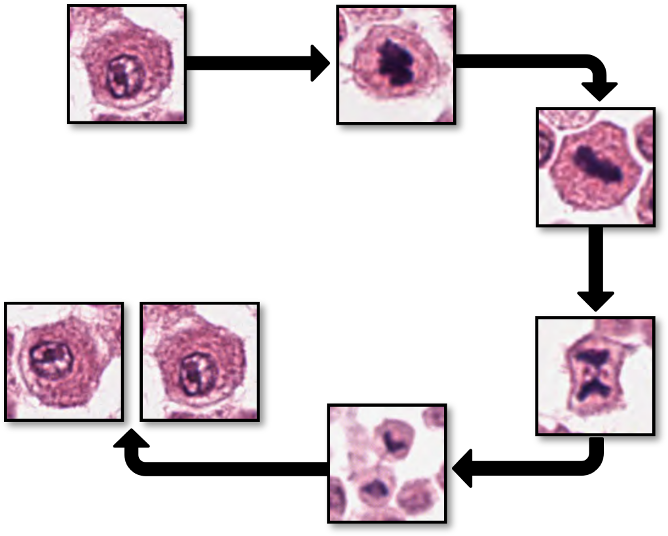
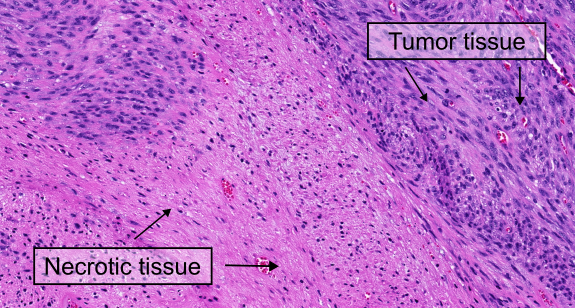
Despite their potential, AI-based systems still have limitations. One major challenge is the great tissue variation throughout large histological images. For example, false positive predictions often occur in non-cancerous tissue or inflamed and necrotic cancer regions. This happens because most algorithms have only been trained with small, carefully selected image parts of the tumor without accounting for the less “perfect” image regions. To be truly useful in real-world medical settings, AI algorithms need to be more robust and work accurately across all parts of a histological image, not just the ideal regions.
What’s New Compared to MIDOG 2022?
The MIDOG 2025 challenge aims at addressing this problem. Task 1 focuses on detecting mitotic figures across all regions of histological images, including challenging areas such as non-tumor regions, inflamed tissue, and necrotic zones. This task addresses a critical gap in previous challenges, which primarily focused on pre-selected hotspot regions. By expanding the scope to whole-slide image applicability, Task 1 aims to test the robustness of algorithms in real-world scenarios.
The Training Dataset(s)
This year’s MIDOG challenge introduces a significant change: for the first time, participants are allowed to use all publicly available datasets to develop robust detection algorithms for Task 1. Over the past few years, several large-scale mitotic figure datasets have been published, offering a wealth of diverse images spanning multiple domains. This opens up new opportunities for participants to leverage a wide range of resources to improve their models. For a list of availabe datasets have a look at our dataset page.
At the heart of this challenge is the MIDOG++ dataset, the most diverse mitotic figure dataset to date. It includes 11,937 mitotic figure annotations across 454 labeled images from 9 distinct domains, which are defined by unique combinations of tumor types, species, scanners, and laboratories. This dataset provides an excellent foundation for developing algorithms capable of robust generalization to unseen domains in the test set.
To address the challenge of detecting mitotic figures across the variability of whole-slide image, including necrotic areas and inflammatory regions, participants can also benefit from fully annotated whole-slide image datasets. For example, the MITOS_WSI_CCMCT dataset contains over 40,000 mitotic figure annotations across 32 fully annotated whole-slide images of canine cutaneous mast cell tumors. While this dataset represents a single domain, it covers a wide variety of tissue types, making it a valuable resource for training algorithms to detect mitotic figures in non-tumor regions or outside traditional tumor hotspots, which are frequently encountered in routine diagnostics.
By combining the diversity of the MIDOG++ dataset with other publicly available datasets, participants have the opportunity to develop algorithms that perform robustly across a wide range of histological contexts, paving the way for real-world applicability.
The Test Dataset(s)
The MIDOG 2025 test set is a carefully curated and diverse dataset designed to push the boundaries of mitotic figure detection algorithms. It represents a significant step forward in evaluating the robustness and generalizability of computational methods in histopathology. With its wide range of tumor types, challenging regions, and realistic variability, the test set provides a rigorous benchmark for assessing algorithm performance in real-world scenarios.
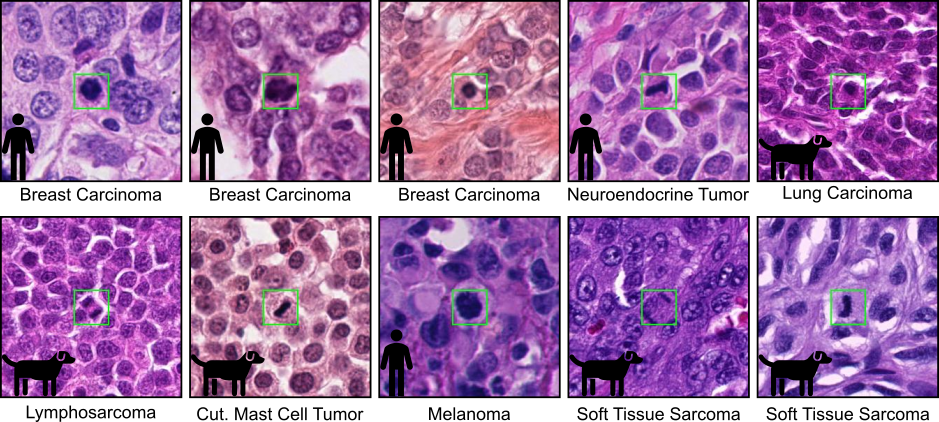
The test set consists of 120 cases, covering 12 distinct tumor types from both human and veterinary pathology. Each tumor type is represented by 10 cases, ensuring a balanced and comprehensive evaluation. These tumor types include well-known examples such as lung adenocarcinoma, glioblastoma, round cell tumors, epithelial tumors, and mesenchymal tumors. To further enhance the diversity of the dataset, two additional tumor types from a new laboratory have been included, expanding the scope beyond what was available in the MIDOG 2022 challenge. This extension ensures that the test set reflects a broader range of histological contexts, making it one of the most diverse datasets ever used in a mitotic figure detection challenge.
One of the most exciting aspects of the MIDOG 2025 test set is its inclusion of regions with varying levels of difficulty. As in previous challenges, the dataset contains regions of interest (ROIs) typically used in tumor grading schemes. However, this year’s test set goes beyond these traditional areas by incorporating random selections from the whole-slide images (WSIs) of the 120 cases. These random regions capture the natural variability of histological slides, providing a more realistic representation of the challenges faced in routine diagnostics. Additionally, the test set includes highly challenging regions, such as areas of inflammation and necrosis. These regions are particularly difficult because they often contain cells with high morphological similarity to mitotic figures, making them prone to false positives. By including these complex areas, the test set ensures that algorithms are tested not only for accuracy but also for their ability to handle the most demanding scenarios.
To support participants during the development phase, a preliminary debug dataset will be made available through the Grand Challenge platform. This dataset includes four tumor types that are not part of the final test set, allowing participants to verify the functionality of their algorithms without revealing the characteristics of the final test cases. The preliminary debug dataset is designed to mimic the overall structure and challenges of the final test set, including random selections and difficult regions such as necrosis and inflammation.
What Kind of Algorithm Do You Need to Develop?
Participants are tasked with developing a mitotic figure detection algorithm that is both accurate and robust, capable of generalizing across diverse tissue types, tumor types, and imaging conditions. The algorithm must handle the inherent variability of large histological images and adapt to unseen domains without relying on prior knowledge of the specific characteristics of the target cases.
Key challenges include detecting mitotic figures not only in traditional tumor hotspots but also across complex regions such as inflamed tissue and necrotic zones, which often contain structures resembling mitotic figures. The algorithm should minimize false positives in these challenging areas while maintaining high sensitivity.
To address these challenges, participants can explore techniques such as domain adaptation, domain-invariant feature extraction, style transfer, test-time augmentation, or ensembling. Creativity is encouraged. Innovative approaches that improve generalization and robustness will be key to success. By tackling these issues, participants will contribute to the development of tools that enhance diagnostic accuracy and reproducibility across laboratories worldwide.
Evaluation metrics
The common metric for evaluation mitotic figure detections is the F1 score and we will use it for evaluation of the submissions in the same way as in previous iterations of MIDOG. The main reason why we will use non-threshold based metrics like mAP is that the perfect detection threshold may vary strongly with the domain, so participants need to optimize a (domain-agnostic) detection threshold.
The F1 metric accounts for false positives as well as false negatives and is thus a fair metric. As some images might have only a small number (or even none at all) mitotic figures (in low grade tumors), we calculate the F1 score over the complete data set by counting all true positives (TP), false positives (FP), and false negatives (FN) and calculate:
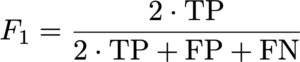
Similarly to previous MIDOG challenges, the F1 score will be calculated over all cases of the test set (i.e. no averaging will be done on a case level).
